Segnale redox antiossidante.2014 gennaio 20;20(3):460-73. doi: 10.1089/ars.2013.5371. Epub 2013 2 agosto.
Katherine R Parzych 1,
Daniel J. Klionsky Affiliazione
- 1Dipartimento di Biologia Molecolare, Cellulare e dello Sviluppo, Istituto di Scienze della Vita, Università del Michigan, Ann Arbor, Michigan.
Articolo PMC gratuito
Astratto
Significato: l'autofagia è un processo di riciclaggio cellulare eucariotico altamente conservato. Attraverso la degradazione di organelli, proteine e macromolecole citoplasmatiche e il riciclaggio dei prodotti di degradazione, l'autofagia svolge un ruolo importante nella sopravvivenza e nel mantenimento delle cellule. Di conseguenza, la disfunzione di questo processo contribuisce alle patologie di molte malattie umane.
Avanzamenti recenti: sono attualmente in corso ricerche approfondite per comprendere meglio il processo di autofagia. In questa recensione, descriviamo l'attuale conoscenza della morfologia, del meccanismo molecolare e della regolazione dell'autofagia dei mammiferi.
Criticità: a livello meccanicistico e normativo, ci sono ancora molte domande senza risposta e punti di confusione che devono ancora essere risolti.
Direzioni future: attraverso ulteriori ricerche, un quadro più completo e accurato del meccanismo molecolare e della regolazione dell'autofagia non solo rafforzerà la nostra comprensione di questo significativo processo cellulare, ma aiuterà nello sviluppo di nuovi trattamenti per le malattie umane in cui l'autofagia non è funzionando correttamente.
FIG. 1. Three types of autophagy in mammalian cells. Macroautophagy relies on de novo formation of cytosolic double-membrane vesicles, autophagosomes, to sequester and transport cargo to the lysosome. Chaperone-mediated autophagy transports individual unfolded proteins directly across the lysosomal membrane. Microautophagy involves the direct uptake of cargo through invagination of the lysosomal membrane. All three types of autophagy lead to degradation of cargo and release of the breakdown products back into the cytosol for reuse by the cell. See the text for details. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
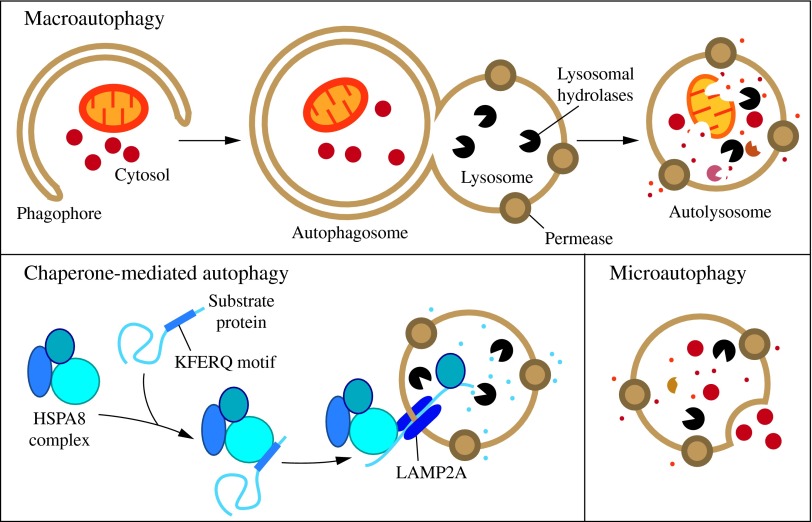 FIG. 2. Morphology of macroautophagy. Nucleation of the phagophore occurs following induction by the ULK1/2 complex. Elongation of the phagophore is aided by the ATG12–ATG5-ATG16L1 complex, the class III PtdIns3K complex, LC3-II, and ATG9. Eventually, the expanding membrane closes around its cargo to form an autophagosome and LC3-II is cleaved from the outer membrane of this structure. The outer membrane of the autophagosome will then fuse with the lysosomal membrane to form an autolysosome. In some instances, the autophagosome may fuse with an endosome, forming an amphisome, before fusing with the lysosome. The contents of the autolysosome are then degraded and exported back into the cytoplasm for reuse by the cell. See the text for details. This figure was modified from Figure 1 in Yang and Klionsky (153). ATG, autophagy-related; PtdIns3K, phosphatidylinositol 3-kinase; ULK, unc-51-like kinase (C. elegans). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 2. Morphology of macroautophagy. Nucleation of the phagophore occurs following induction by the ULK1/2 complex. Elongation of the phagophore is aided by the ATG12–ATG5-ATG16L1 complex, the class III PtdIns3K complex, LC3-II, and ATG9. Eventually, the expanding membrane closes around its cargo to form an autophagosome and LC3-II is cleaved from the outer membrane of this structure. The outer membrane of the autophagosome will then fuse with the lysosomal membrane to form an autolysosome. In some instances, the autophagosome may fuse with an endosome, forming an amphisome, before fusing with the lysosome. The contents of the autolysosome are then degraded and exported back into the cytoplasm for reuse by the cell. See the text for details. This figure was modified from Figure 1 in Yang and Klionsky (153). ATG, autophagy-related; PtdIns3K, phosphatidylinositol 3-kinase; ULK, unc-51-like kinase (C. elegans). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
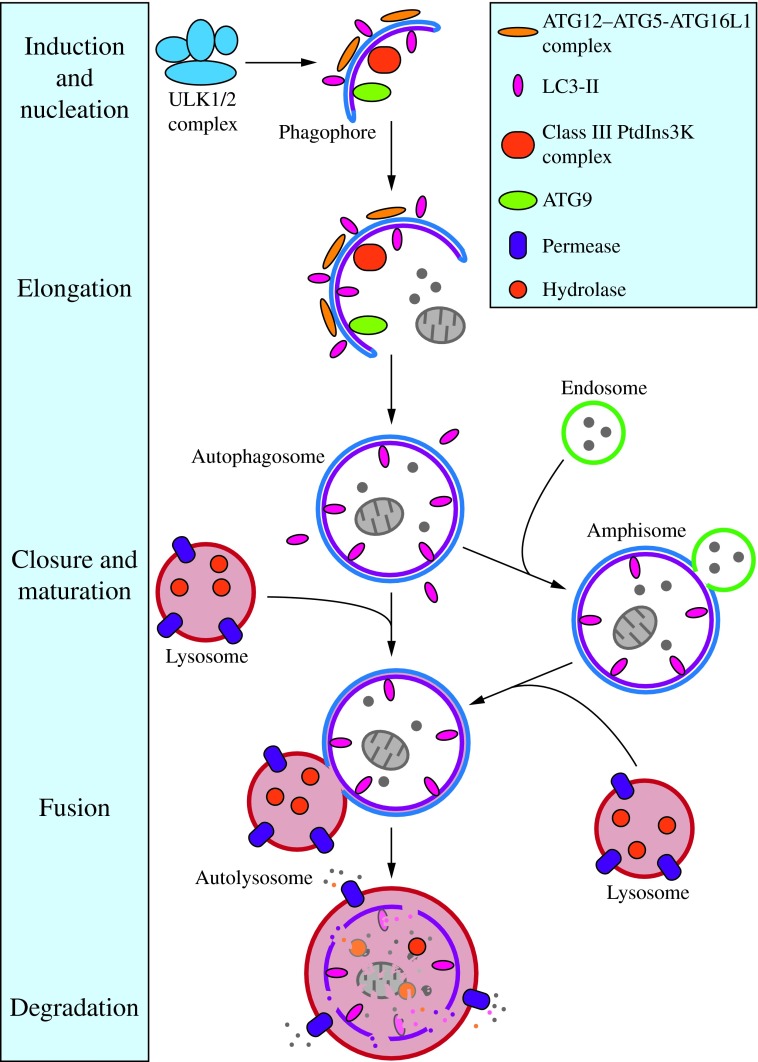 FIG. 3. The induction complex consists of ULK1/2, ATG13, RB1CC1, and C12orf44. Under nutrient-rich conditions, MTORC1 associates with the complex and inactivates ULK1/2 and ATG13 through phosphorylation. During starvation, MTORC1 dissociates from the complex and ATG13 and ULK1/2 become partially dephosphorylated by as yet unidentified phosphatases, allowing the complex to induce macroautophagy. RB1CC1/FIP200 and C12orf44/ATG101 are also associated with the induction complex and are essential for macroautophagy. RB1CC1/FIP200 may be the ortholog of yeast Atg17, whereas the function of C12orf44/ATG101 is not known. This figure was modified from Figure 1 in Yang and Klionsky (154). MTORC1, mechanistic target of rapamycin complex 1; RB1CC1, RB1-inducible coiled-coil 1. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 3. The induction complex consists of ULK1/2, ATG13, RB1CC1, and C12orf44. Under nutrient-rich conditions, MTORC1 associates with the complex and inactivates ULK1/2 and ATG13 through phosphorylation. During starvation, MTORC1 dissociates from the complex and ATG13 and ULK1/2 become partially dephosphorylated by as yet unidentified phosphatases, allowing the complex to induce macroautophagy. RB1CC1/FIP200 and C12orf44/ATG101 are also associated with the induction complex and are essential for macroautophagy. RB1CC1/FIP200 may be the ortholog of yeast Atg17, whereas the function of C12orf44/ATG101 is not known. This figure was modified from Figure 1 in Yang and Klionsky (154). MTORC1, mechanistic target of rapamycin complex 1; RB1CC1, RB1-inducible coiled-coil 1. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
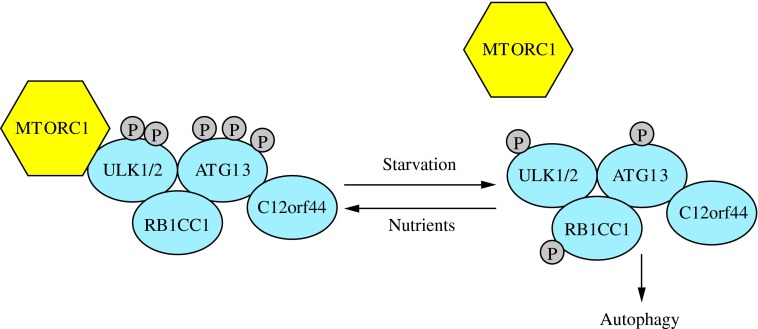 FIG. 4. The activity of the class III PtdIns3K complex is regulated by subunit composition. The ATG14 complex (ATG14-BECN1-PIK3C3-PIK3R4) is required for macroautophagy. It can be positively regulated by AMBRA1 and negatively regulated by BCL2 binding to BECN1 and preventing association with the complex. The UVRAG (UVRAG-BECN1-PIK3C3-PIK3R4) complex is involved in the endocytic pathway and also participates in macroautophagy. SH3GLB1/Bif-1 positively regulates this complex by binding UVRAG. The KIAA0226/Rubicon complex (KIAA0226-UVRAG-BECN1-PIK3C3-PIK3R4) negatively regulates macroautophagy. This figure was modified from Figure 1 in Yang and Klionsky (154). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 4. The activity of the class III PtdIns3K complex is regulated by subunit composition. The ATG14 complex (ATG14-BECN1-PIK3C3-PIK3R4) is required for macroautophagy. It can be positively regulated by AMBRA1 and negatively regulated by BCL2 binding to BECN1 and preventing association with the complex. The UVRAG (UVRAG-BECN1-PIK3C3-PIK3R4) complex is involved in the endocytic pathway and also participates in macroautophagy. SH3GLB1/Bif-1 positively regulates this complex by binding UVRAG. The KIAA0226/Rubicon complex (KIAA0226-UVRAG-BECN1-PIK3C3-PIK3R4) negatively regulates macroautophagy. This figure was modified from Figure 1 in Yang and Klionsky (154). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
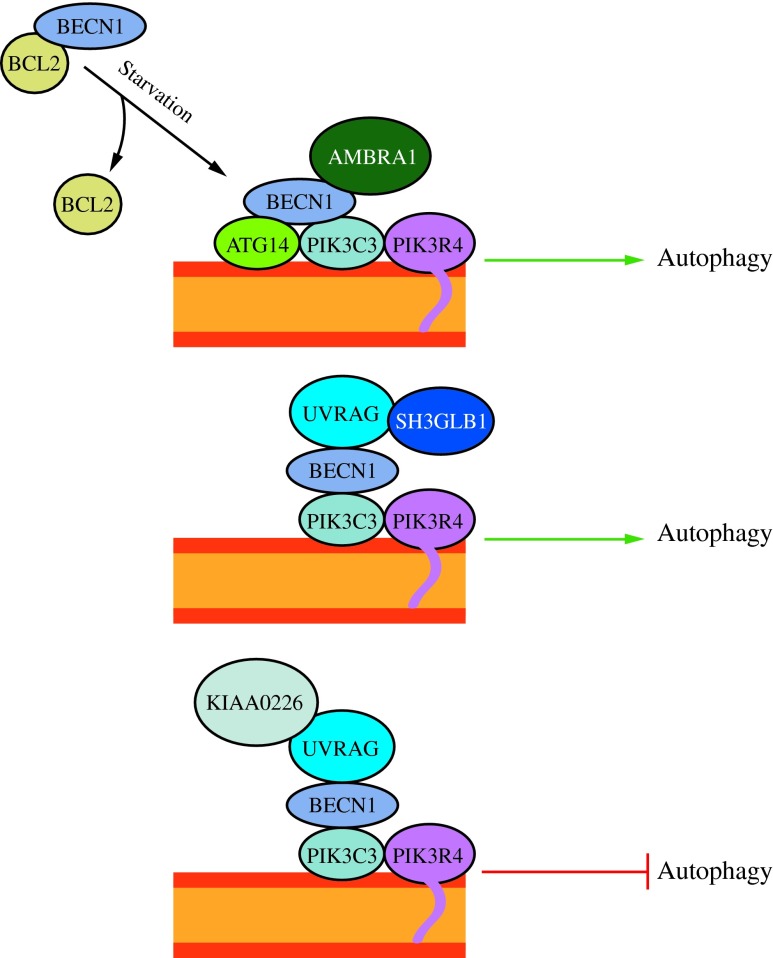 FIG. 5. ATG12–ATG5-ATG16L1 conjugation complex. The ubiquitin-like protein ATG12 is irreversibly conjugated to ATG5 in an ATG7- and ATG10-dependent manner. ATG7 and ATG10 function as E1 activating and E2 conjugating enzymes, respectively. The ATG12–ATG5 conjugate binds ATG16L1 through ATG5. ATG16L1 dimerizes and allows association with the phagophore, promoting membrane expansion. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 5. ATG12–ATG5-ATG16L1 conjugation complex. The ubiquitin-like protein ATG12 is irreversibly conjugated to ATG5 in an ATG7- and ATG10-dependent manner. ATG7 and ATG10 function as E1 activating and E2 conjugating enzymes, respectively. The ATG12–ATG5 conjugate binds ATG16L1 through ATG5. ATG16L1 dimerizes and allows association with the phagophore, promoting membrane expansion. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
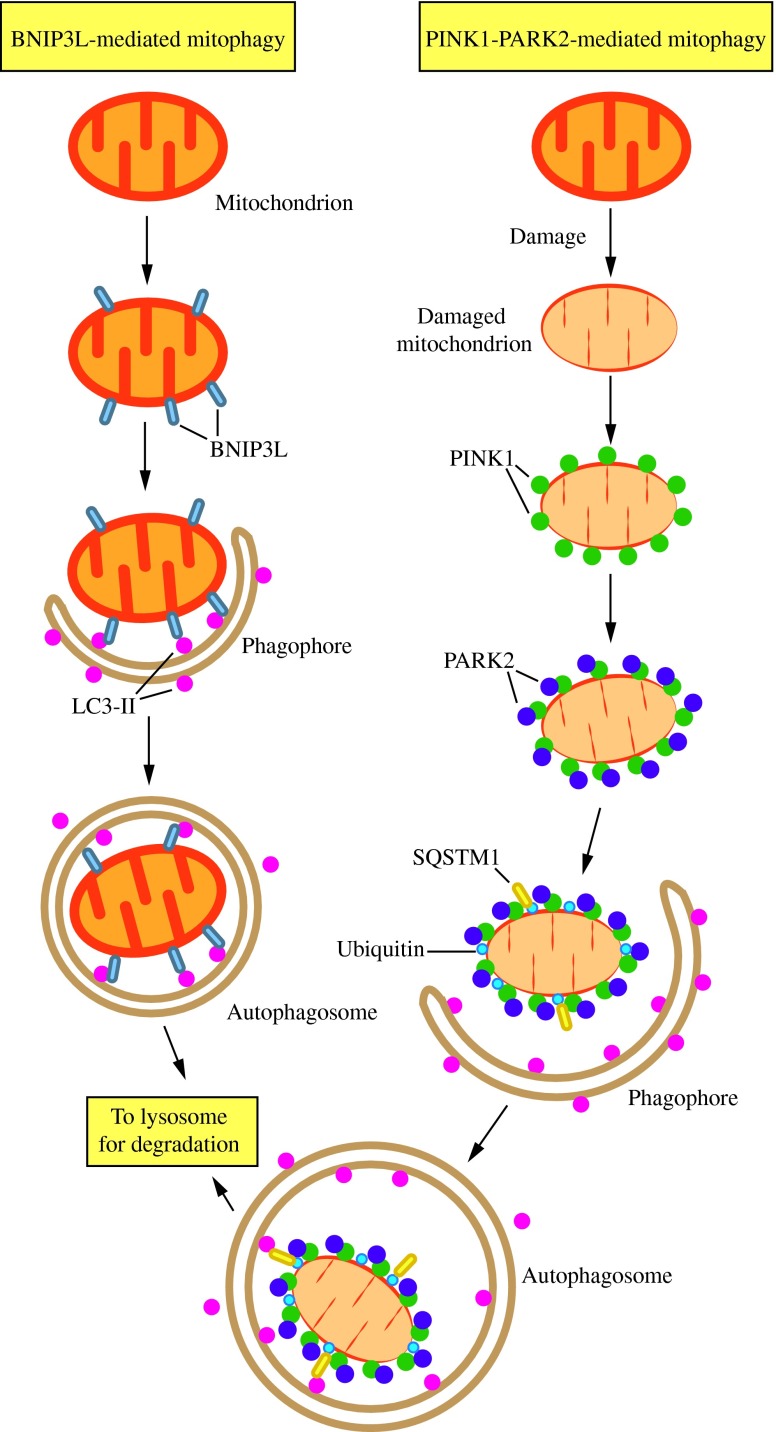
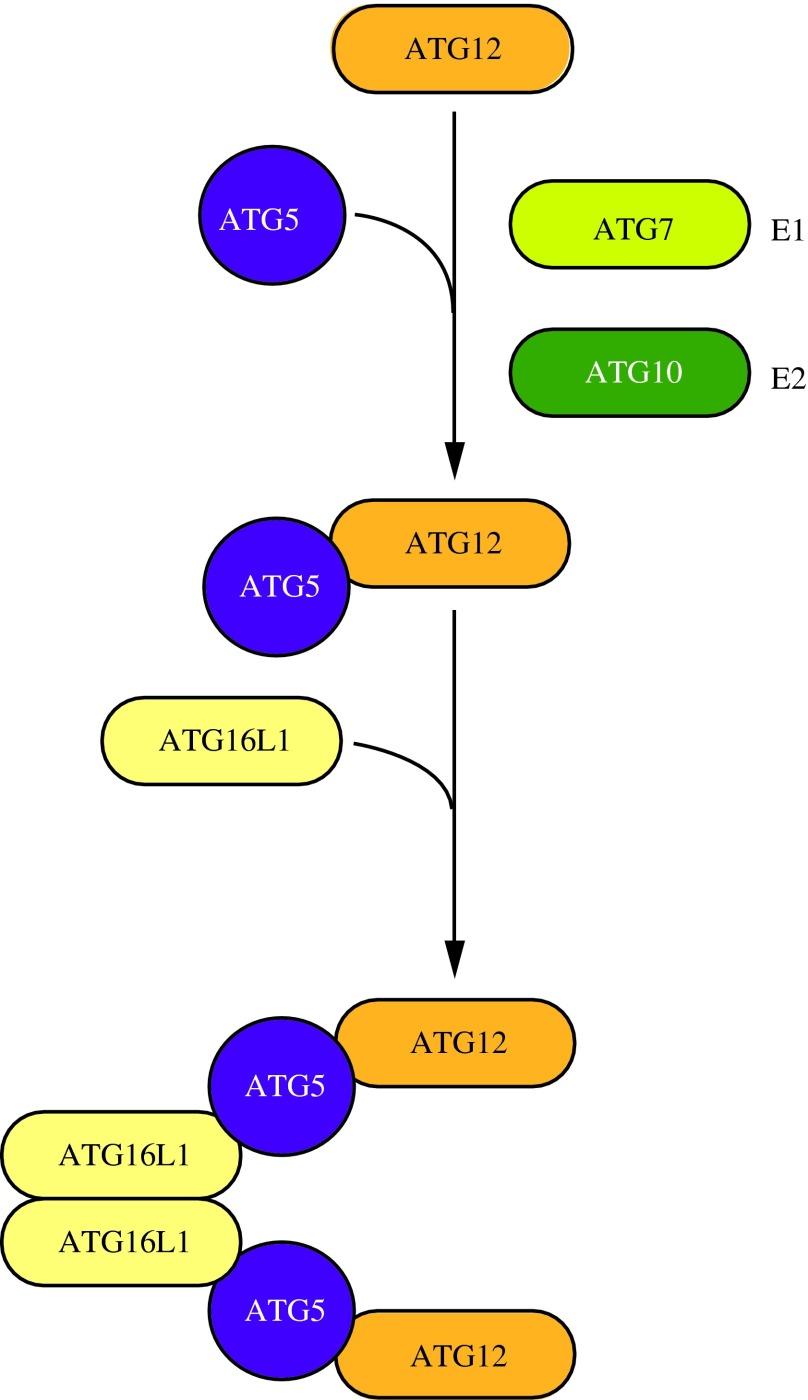 FIG. 6. Two mechanisms of mitophagy. Mitochondria are cleared from maturing red blood cells through a mechanism involving autophagic recognition of mitochondria through a BNIP3L–LC3 interaction. During removal of damaged mitochondria, PARK2 binds to PINK1 on the mitochondrial surface and ubiquitinates mitochondrial outer membrane proteins, which may then bind SQSTM1, a receptor that interacts with LC3. In either case, the interaction with LC3 leads to sequestration by the phagophore and eventual degradation. This figure was modified from Figure 2 of Youle and Narendra (158). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 6. Two mechanisms of mitophagy. Mitochondria are cleared from maturing red blood cells through a mechanism involving autophagic recognition of mitochondria through a BNIP3L–LC3 interaction. During removal of damaged mitochondria, PARK2 binds to PINK1 on the mitochondrial surface and ubiquitinates mitochondrial outer membrane proteins, which may then bind SQSTM1, a receptor that interacts with LC3. In either case, the interaction with LC3 leads to sequestration by the phagophore and eventual degradation. This figure was modified from Figure 2 of Youle and Narendra (158). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars